Irina Voiculescu
Reg4Pru: Regularisation Through Random Token Routing for Token Pruning
Feb 03, 2026Abstract:Transformers are widely adopted in modern vision models due to their strong ability to scale with dataset size and generalisability. However, this comes with a major drawback: computation scales quadratically to the total number of tokens. Numerous methods have been proposed to mitigate this. For example, we consider token pruning with reactivating tokens from preserved representations, but the increased computational efficiency of this method results in decreased stability from the preserved representations, leading to poorer dense prediction performance at deeper layers. In this work, we introduce Reg4Pru, a training regularisation technique that mitigates token-pruning performance loss for segmentation. We compare our models on the FIVES blood vessel segmentation dataset and find that Reg4Pru improves average precision by an absolute 46% compared to the same model trained without routing. This increase is observed using a configuration that achieves a 29% relative speedup in wall-clock time compared to the non-pruned baseline. These findings indicate that Reg4Pru is a valuable regulariser for token reduction strategies.
Automated Landmark Detection for assessing hip conditions: A Cross-Modality Validation of MRI versus X-ray
Jan 26, 2026Abstract:Many clinical screening decisions are based on angle measurements. In particular, FemoroAcetabular Impingement (FAI) screening relies on angles traditionally measured on X-rays. However, assessing the height and span of the impingement area requires also a 3D view through an MRI scan. The two modalities inform the surgeon on different aspects of the condition. In this work, we conduct a matched-cohort validation study (89 patients, paired MRI/X-ray) using standard heatmap regression architectures to assess cross-modality clinical equivalence. Seen that landmark detection has been proven effective on X-rays, we show that MRI also achieves equivalent localisation and diagnostic accuracy for cam-type impingement. Our method demonstrates clinical feasibility for FAI assessment in coronal views of 3D MRI volumes, opening the possibility for volumetric analysis through placing further landmarks. These results support integrating automated FAI assessment into routine MRI workflows. Code is released at https://github.com/Malga-Vision/Landmarks-Hip-Conditions
MUPAX: Multidimensional Problem Agnostic eXplainable AI
Jul 17, 2025Abstract:Robust XAI techniques should ideally be simultaneously deterministic, model agnostic, and guaranteed to converge. We propose MULTIDIMENSIONAL PROBLEM AGNOSTIC EXPLAINABLE AI (MUPAX), a deterministic, model agnostic explainability technique, with guaranteed convergency. MUPAX measure theoretic formulation gives principled feature importance attribution through structured perturbation analysis that discovers inherent input patterns and eliminates spurious relationships. We evaluate MUPAX on an extensive range of data modalities and tasks: audio classification (1D), image classification (2D), volumetric medical image analysis (3D), and anatomical landmark detection, demonstrating dimension agnostic effectiveness. The rigorous convergence guarantees extend to any loss function and arbitrary dimensions, making MUPAX applicable to virtually any problem context for AI. By contrast with other XAI methods that typically decrease performance when masking, MUPAX not only preserves but actually enhances model accuracy by capturing only the most important patterns of the original data. Extensive benchmarking against the state of the XAI art demonstrates MUPAX ability to generate precise, consistent and understandable explanations, a crucial step towards explainable and trustworthy AI systems. The source code will be released upon publication.
Entropy Bootstrapping for Weakly Supervised Nuclei Detection
Nov 20, 2024



Abstract:Microscopy structure segmentation, such as detecting cells or nuclei, generally requires a human to draw a ground truth contour around each instance. Weakly supervised approaches (e.g. consisting of only single point labels) have the potential to reduce this workload significantly. Our approach uses individual point labels for an entropy estimation to approximate an underlying distribution of cell pixels. We infer full cell masks from this distribution, and use Mask-RCNN to produce an instance segmentation output. We compare this point--annotated approach with training on the full ground truth masks. We show that our method achieves a comparatively good level of performance, despite a 95% reduction in pixel labels.
Parallel Watershed Partitioning: GPU-Based Hierarchical Image Segmentation
Oct 11, 2024



Abstract:Many image processing applications rely on partitioning an image into disjoint regions whose pixels are 'similar.' The watershed and waterfall transforms are established mathematical morphology pixel clustering techniques. They are both relevant to modern applications where groups of pixels are to be decided upon in one go, or where adjacency information is relevant. We introduce three new parallel partitioning algorithms for GPUs. By repeatedly applying watershed algorithms, we produce waterfall results which form a hierarchy of partition regions over an input image. Our watershed algorithms attain competitive execution times in both 2D and 3D, processing an 800 megavoxel image in less than 1.4 sec. We also show how to use this fully deterministic image partitioning as a pre-processing step to machine learning based semantic segmentation. This replaces the role of superpixel algorithms, and results in comparable accuracy and faster training times.
Optimising for the Unknown: Domain Alignment for Cephalometric Landmark Detection
Oct 06, 2024Abstract:Cephalometric Landmark Detection is the process of identifying key areas for cephalometry. Each landmark is a single GT point labelled by a clinician. A machine learning model predicts the probability locus of a landmark represented by a heatmap. This work, for the 2024 CL-Detection MICCAI Challenge, proposes a domain alignment strategy with a regional facial extraction module and an X-ray artefact augmentation procedure. The challenge ranks our method's results as the best in MRE of 1.186mm and third in the 2mm SDR of 82.04% on the online validation leaderboard. The code is available at https://github.com/Julian-Wyatt/OptimisingfortheUnknown.
Salt & Pepper Heatmaps: Diffusion-informed Landmark Detection Strategy
Jul 12, 2024



Abstract:Anatomical Landmark Detection is the process of identifying key areas of an image for clinical measurements. Each landmark is a single ground truth point labelled by a clinician. A machine learning model predicts the locus of a landmark as a probability region represented by a heatmap. Diffusion models have increased in popularity for generative modelling due to their high quality sampling and mode coverage, leading to their adoption in medical image processing for semantic segmentation. Diffusion modelling can be further adapted to learn a distribution over landmarks. The stochastic nature of diffusion models captures fluctuations in the landmark prediction, which we leverage by blurring into meaningful probability regions. In this paper, we reformulate automatic Anatomical Landmark Detection as a precise generative modelling task, producing a few-hot pixel heatmap. Our method achieves state-of-the-art MRE and comparable SDR performance with existing work.
Runtime Freezing: Dynamic Class Loss for Multi-Organ 3D Segmentation
Jun 12, 2024

Abstract:Segmentation has become a crucial pre-processing step to many refined downstream tasks, and particularly so in the medical domain. Even with recent improvements in segmentation models, many segmentation tasks remain difficult. When multiple organs are segmented simultaneously, difficulties are due not only to the limited availability of labelled data, but also to class imbalance. In this work we propose dynamic class-based loss strategies to mitigate the effects of highly imbalanced training data. We show how our approach improves segmentation performance on a challenging Multi-Class 3D Abdominal Organ dataset.
Infant hip screening using multi-class ultrasound scan segmentation
Nov 08, 2022



Abstract:Developmental dysplasia of the hip (DDH) is a condition in infants where the femoral head is incorrectly located in the hip joint. We propose a deep learning algorithm for segmenting key structures within ultrasound images, employing this to calculate Femoral Head Coverage (FHC) and provide a screening diagnosis for DDH. To our knowledge, this is the first study to automate FHC calculation for DDH screening. Our algorithm outperforms the international state of the art, agreeing with expert clinicians on 89.8% of our test images.
Triple-View Feature Learning for Medical Image Segmentation
Aug 12, 2022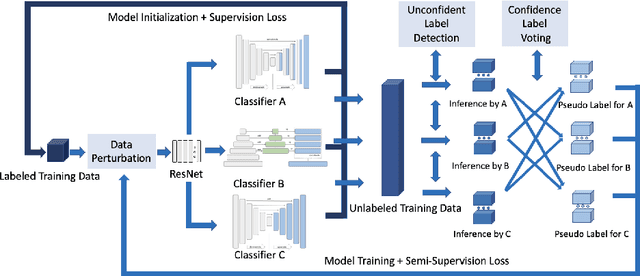
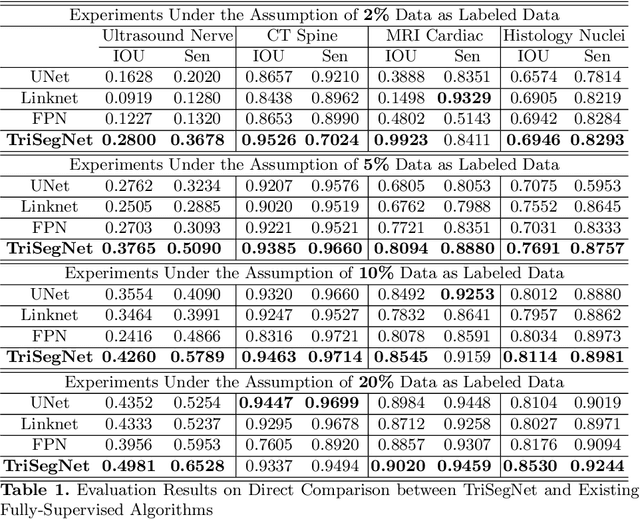
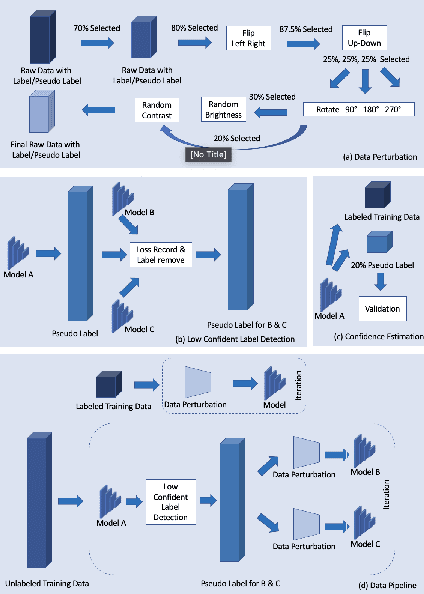
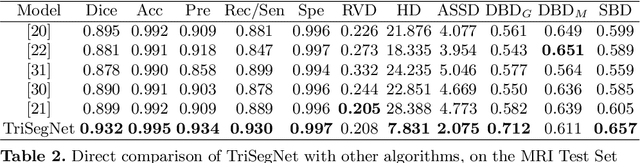
Abstract:Deep learning models, e.g. supervised Encoder-Decoder style networks, exhibit promising performance in medical image segmentation, but come with a high labelling cost. We propose TriSegNet, a semi-supervised semantic segmentation framework. It uses triple-view feature learning on a limited amount of labelled data and a large amount of unlabeled data. The triple-view architecture consists of three pixel-level classifiers and a low-level shared-weight learning module. The model is first initialized with labelled data. Label processing, including data perturbation, confidence label voting and unconfident label detection for annotation, enables the model to train on labelled and unlabeled data simultaneously. The confidence of each model gets improved through the other two views of the feature learning. This process is repeated until each model reaches the same confidence level as its counterparts. This strategy enables triple-view learning of generic medical image datasets. Bespoke overlap-based and boundary-based loss functions are tailored to the different stages of the training. The segmentation results are evaluated on four publicly available benchmark datasets including Ultrasound, CT, MRI, and Histology images. Repeated experiments demonstrate the effectiveness of the proposed network compared against other semi-supervised algorithms, across a large set of evaluation measures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge